ICA
Introductory notes:
This notebook presents ICA artifact repairment functionality:
Fitting ICA
Exploring components
Excluding components from the raw data
Recommended reading:
Import data
Import module
from sleepeegpy.pipeline import ICAPipe
from os import makedirs, path
import mne
mne.set_log_level("ERROR")
Initialize ICAPipe object
If you wish to change the path for output_dir ot input dir, change it below. If no such folders, they will be created automatically.
output_dir = "output_folder" # Output path and name can be changed here
input_dir = "input_files" # input files dir can be changed here
makedirs(input_dir, exist_ok=True)
makedirs(output_dir, exist_ok=True)
Add required files
Put all your files in the input folder.
Modify your eeg file name below. The file can be any format supported by the mne.read_raw() function.
For more information about the supported formats, see mne documentation
eeg_file_name = "resampled_raw.fif"
ica_file_name = (
None # If you want to load already fitted ICA - replace None with the file name.
)
path_to_eeg = path.join(input_dir, eeg_file_name)
path_to_ica = path.join(input_dir, ica_file_name) if ica_file_name else None
ica_pipe = ICAPipe(
# can be any type of eeg file that MNE's read_raw() function supports.
path_to_eeg=path_to_eeg,
output_dir=output_dir,
method="fastica", # ICA method, can be either 'fastica', 'infomax' or 'picard'
fit_params=None, # Additional parameters passed to the ICA estimator as specified by 'method'.
n_components=40, # was 40, Number of principal components that are passed to the ICA algorithm during fitting.
random_state=120, # A seed for the NumPy random number generator (RNG).
path_to_ica=path_to_ica,
)
Fit
Run the ICA decomposition on 1 Hz high-pass filtered data
ica_pipe.fit(filter_kwargs=dict(l_freq=1.0, h_freq=None, n_jobs=-1))
Explore
Plot independent components from raw data, you can select here components you want to exclude.
ica_pipe.plot_sources()
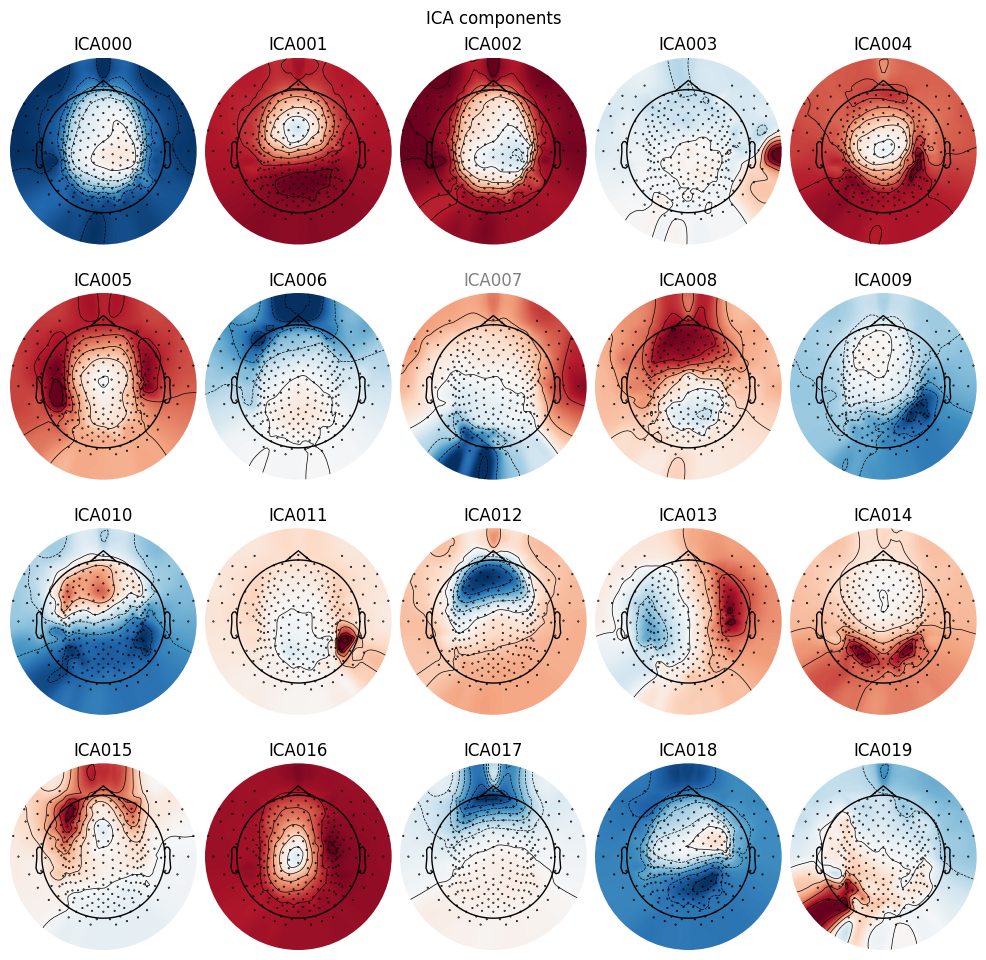
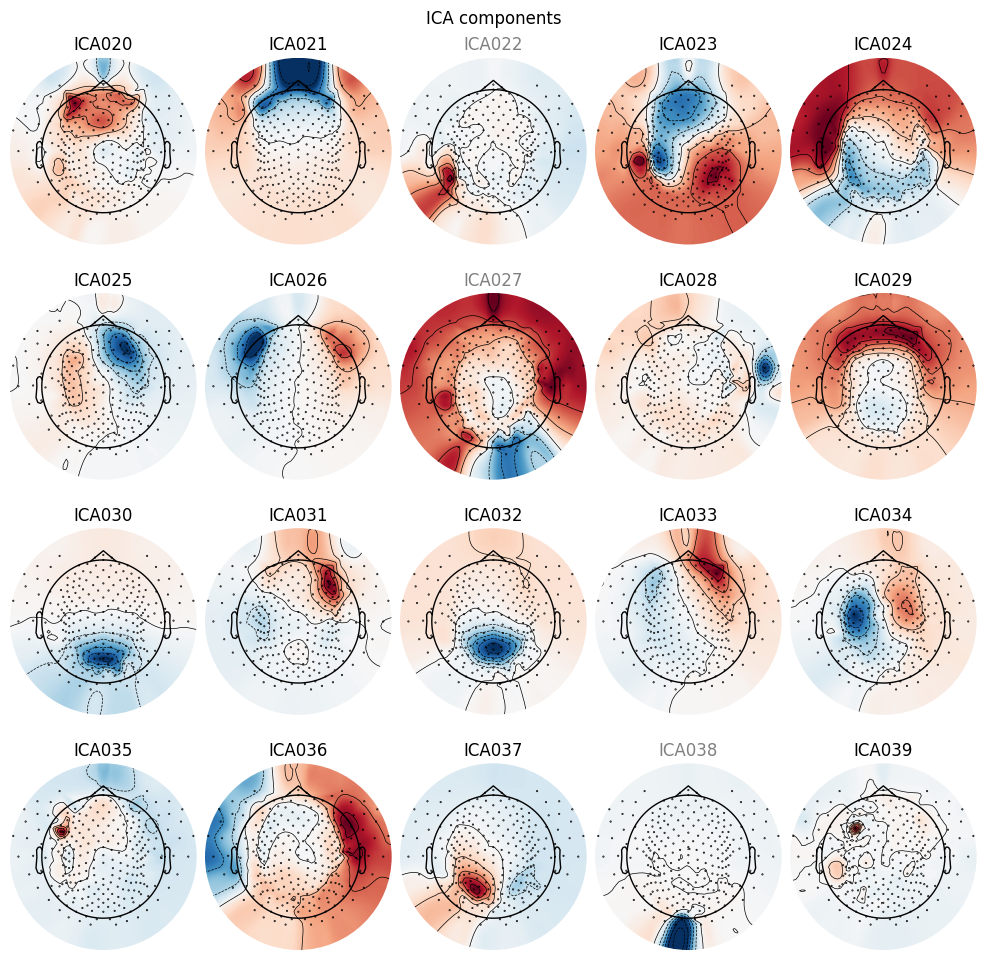
Plot components on interpolated sensor topography.
_ = ica_pipe.plot_components()
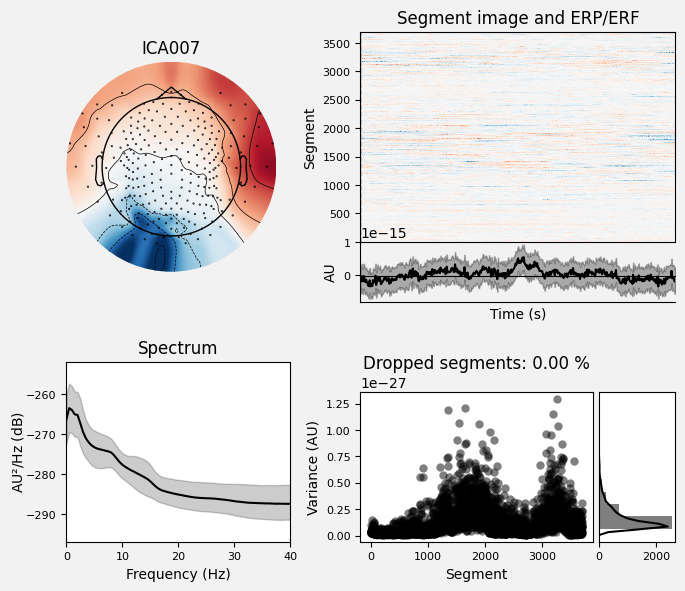
Display component properties.
_ = ica_pipe.plot_properties(picks=[7], psd_args=dict(fmin=0, fmax=40))
Exclude
Apply to the raw signal ICA component exclusion.
ica_pipe.apply()
Save repaired EEG signal and ica solution
ica_pipe.save_raw("after_ica_raw.fif", overwrite=True)
ica_pipe.save_ica(fname="exclude-ica.fif", overwrite=True)