Spindles analysis
Introductory notes:
This notebook presents spindles detection and analysis functionality.
Recommended reading:
Import module
from sleepeegpy.pipeline import SpindlesPipe
from joblib import parallel_backend
parallel_backend("loky", n_jobs=-1)
from os import makedirs, path
Spindles detection
If you wish to change the path for output_dir ot input dir, change it below. If no such folders, they will be created automatically.
output_dir = "output_folder" # Output path and name can be changed here
input_dir = "input_files" # input files dir can be changed here
makedirs(input_dir, exist_ok=True)
makedirs(output_dir, exist_ok=True)
Add required files and data
Put all your files in the input folder.
Modify your eeg file name below. The file can be any format supported by the mne.read_raw() function.
Modify your hypnogram file name below
Make sure the hypno_freq is the right frequency.
For more information about the supported formats, see mne documentation
eeg_file_name = "resampled_raw.fif" # add your eeg_path here
hypnogram_filename = "staging.txt" # Hypnogram filename can be changed here (file must be in the input dir)
hypno_freq = 1
path_to_eeg = path.join(input_dir, eeg_file_name)
hypnogram_path = path.join(input_dir, hypnogram_filename)
spindles_pipe = SpindlesPipe(
path_to_eeg=path_to_eeg,
output_dir=output_dir,
path_to_hypno=hypnogram_path,
hypno_freq=hypno_freq,
)
Please refer to the YASA’s documentation for details about the arguments.
spindles_pipe.detect(
picks=["eeg"],
reference="average",
include=(2, 3),
freq_sp=(12, 15),
freq_broad=(1, 30),
duration=(0.5, 2),
min_distance=500,
thresh={"corr": 0.65, "rel_pow": 0.2, "rms": 1.5},
multi_only=False,
remove_outliers=False,
verbose=False,
save=True,
)
Please refer to the YASA’s documentation for details about the arguments.
spindles_pipe.results.summary(
grp_chan=False, grp_stage=True, mask=None, aggfunc="mean", sort=True
)
| Count | Density | Duration | Amplitude | RMS | AbsPower | RelPower | Frequency | Oscillations | Symmetry | |
|---|---|---|---|---|---|---|---|---|---|---|
| Stage | ||||||||||
| 2 | 48125 | 439.497717 | 0.802510 | 35.844591 | 7.849986 | 1.739719 | 0.402655 | 13.149206 | 10.165049 | 0.513328 |
| 3 | 22294 | 116.722513 | 0.751883 | 40.712861 | 9.045872 | 1.862345 | 0.384414 | 13.194922 | 9.552705 | 0.516585 |
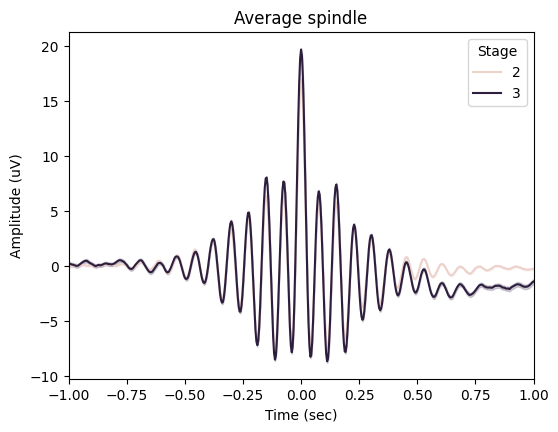
spindles_pipe.plot_average(
save=True,
center="Peak",
hue="Stage",
time_before=1,
time_after=1,
filt=(None, None),
mask=None,
)
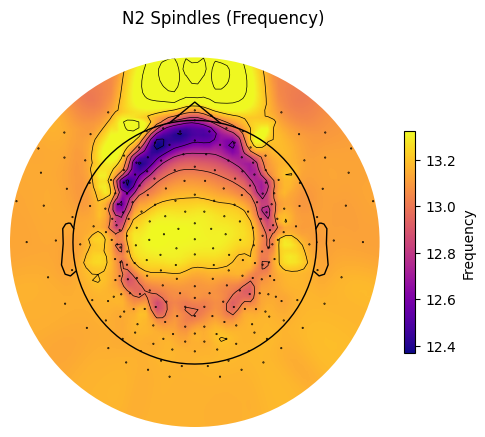
spindles_pipe.plot_topomap(
prop="Frequency", # Any of the event properties returned by pipe.results.summary().
stage="N2", # Stage to plot the topomap for.
aggfunc="mean", # Aggregation function for channel grouping.
# Should contain at least index of the provided "stage".
sleep_stages={"Wake": 0, "N1": 1, "N2": 2, "N3": 3, "REM": 4},
axis=None, # Whether to plot on provided matplotlib axis.
save=True, # Whether to save the plot as a file.
topomap_args=dict(cmap="plasma"), # Arguments passed to mne.viz.plot_topomap().
cbar_args=None, # Arguments passed to plt.colorbar().
)
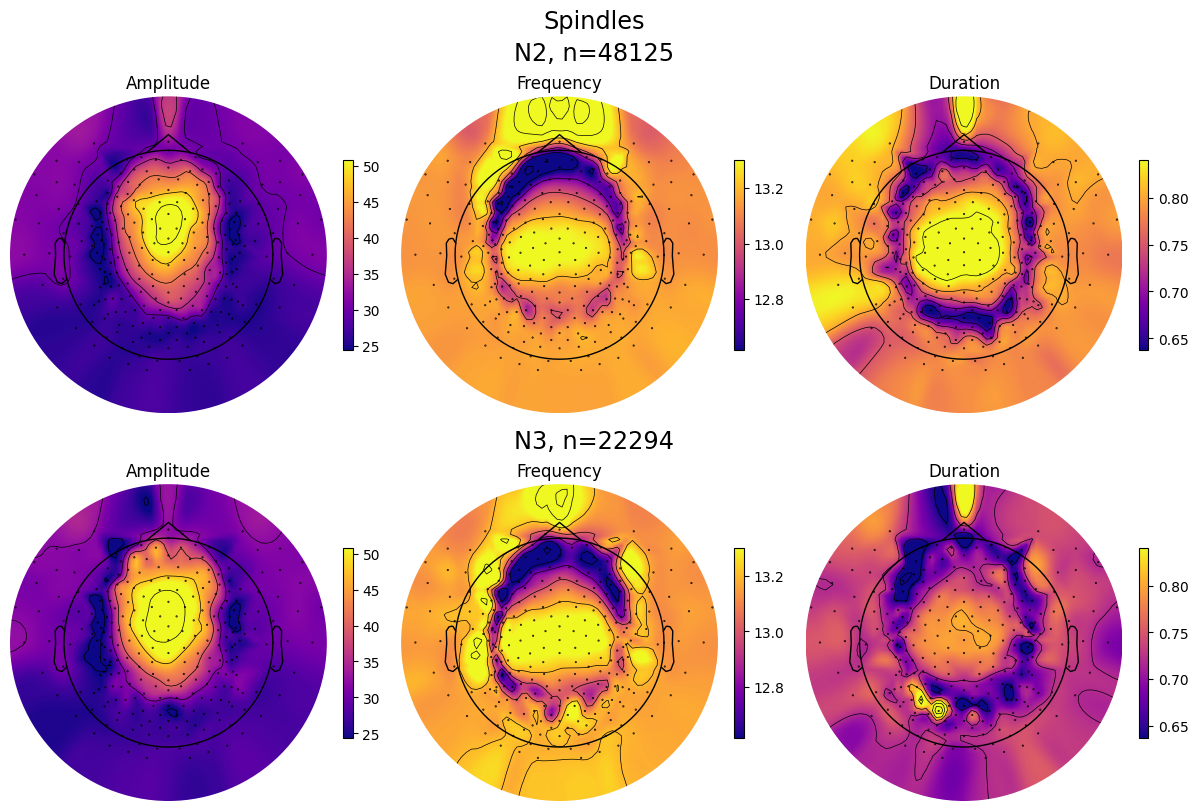
spindles_pipe.plot_topomap_collage(
# Some of the event properties returned by pipe.results.summary().
props=["Amplitude", "Frequency", "Duration"],
aggfunc="mean", # Aggregation function for channel grouping.
# Tuple of strs or "all", e.g., ("N1", "REM") or "all" (plots all "sleep_stages").
stages_to_plot="all",
# Should contain at least indices of the provided "stages_to_plot".
sleep_stages={"N2": 2, "N3": 3},
low_percentile=5, # Set min color value by percentile of the property data.
high_percentile=95, # Set max color value by percentile of the property data.
fig=None, # Instance of plt.Figure, a new fig will be created if None.
save=True, # Whether to save the plot as a file.
topomap_args=dict(cmap="plasma"), # Arguments passed to mne.viz.plot_topomap().
cbar_args=None, # Arguments passed to plt.colorbar().
)
Constructs MNE AverageTFR object for every available sleep stage.
spindles_pipe.compute_tfr(
freqs=(10, 20), # Interval of frequencies of interest.
n_freqs=100, # TFR frequency resolution.
time_before=1, # Time in sec before the peak to use in TFR computation. Should be positive.
time_after=1, # Time in sec after the peak to use in TFR computation.
method="morlet", # TFR method: 'morlet' or 'multitaper'.
save=True, # Whether to save AverageTFR object per sleep stage as hdf5 file.
overwrite=False, # Whether to overwrite existing files.
)
spindles_pipe.tfrs
spindles_pipe.tfrs["N2"].data
array([[[165.44385771, 169.92036454, 174.23956786, ..., 251.72769742,
245.62927484, 239.30345075],
[163.99052483, 168.49114899, 172.83396985, ..., 247.9999998 ,
241.99269192, 235.75219132],
[162.56633576, 167.09405193, 171.46393358, ..., 244.37907057,
238.45905738, 232.30020575],
...,
[ 46.16793334, 47.01757305, 47.5113704 , ..., 63.95819156,
63.84737753, 63.19868073],
[ 45.74479396, 46.57959889, 47.0574021 , ..., 63.33866633,
63.24374934, 62.61033302],
[ 45.3302599 , 46.15072422, 46.61304951, ..., 62.73090346,
62.65140487, 62.03282812]],
[[183.03852542, 188.05132799, 192.90103697, ..., 362.26134796,
352.15546474, 341.75556073],
[179.36273518, 184.3038714 , 189.08251403, ..., 354.76061295,
344.79643891, 334.54188686],
[175.76023984, 180.63134178, 185.34100882, ..., 347.22249221,
337.41589585, 327.32177991],
...,
[ 64.80923984, 65.85678097, 66.37301043, ..., 68.02721018,
67.91046883, 67.22764763],
[ 64.33684292, 65.36606319, 65.86121262, ..., 67.25489205,
67.1579677 , 66.49524398],
[ 63.86404637, 64.87444836, 65.34807822, ..., 66.50109653,
66.42300215, 65.77945011]],
[[357.24759613, 368.793126 , 380.08658706, ..., 588.0948437 ,
572.42633713, 556.26896238],
[347.83432458, 359.13567367, 370.18398281, ..., 583.11395626,
567.25240803, 550.91532737],
[338.32955624, 349.36741722, 360.15064435, ..., 577.22434197,
561.2249059 , 544.76243092],
...,
[ 65.51413465, 66.88240809, 67.8213576 , ..., 89.48868791,
90.12620337, 90.01072393],
[ 65.18081622, 66.53743615, 67.45993565, ..., 88.95798722,
89.60549199, 89.49516761],
[ 64.83682909, 66.17995034, 67.0840514 , ..., 88.45008017,
89.1058455 , 88.99904655]],
...,
[[169.34408048, 175.25910732, 181.08300933, ..., 288.11138222,
278.18552574, 268.18170021],
[165.91686525, 171.72118948, 177.4298448 , ..., 282.09107422,
272.31825781, 262.46583089],
[162.6619232 , 168.36491613, 173.96814788, ..., 276.14859022,
266.53102087, 256.83215105],
...,
[ 33.30850643, 33.94177284, 34.31373952, ..., 40.33239391,
40.13736805, 39.62441846],
[ 32.99837925, 33.6212056 , 33.98223375, ..., 39.79041469,
39.61710556, 39.12574162],
[ 32.69783405, 33.31092644, 33.66175045, ..., 39.26461546,
39.112192 , 38.64157834]],
[[137.05899418, 142.02440564, 146.9221348 , ..., 230.90050237,
223.33624383, 215.66500415],
[133.75877594, 138.62616574, 143.42396777, ..., 227.3662423 ,
219.85924288, 212.2437449 ],
[130.59154621, 135.36711042, 140.07158921, ..., 223.84530067,
216.39593257, 208.83683256],
...,
[ 23.97646908, 24.50242238, 24.84852637, ..., 33.73008036,
33.47365102, 32.95415897],
[ 23.77058193, 24.28991435, 24.62891084, ..., 33.32859906,
33.08517034, 32.57863308],
[ 23.57007977, 24.08317241, 24.4154638 , ..., 32.93668107,
32.70590645, 32.21196209]],
[[431.55839651, 445.42635453, 459.07103265, ..., 600.42672536,
582.1431401 , 563.5192219 ],
[426.92557365, 440.7304316 , 454.30294034, ..., 592.32535875,
574.12609719, 555.58617416],
[422.63123618, 436.39371621, 449.91558109, ..., 584.31384353,
566.1959117 , 547.7378584 ],
...,
[113.55675603, 117.02377124, 119.81135006, ..., 135.2187291 ,
133.06098679, 129.97487896],
[112.32289575, 115.73295768, 118.46154285, ..., 133.76381278,
131.67057141, 128.64501669],
[111.122582 , 114.47794916, 117.14985745, ..., 132.33726145,
130.30705071, 127.34066112]]])
spindles_pipe.tfrs["N2"].to_data_frame()
| time | freq | E1 | E10 | E100 | E101 | E102 | E103 | E104 | E105 | ... | E91 | E92 | E93 | E94 | E95 | E96 | E97 | E98 | E99 | VREF | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | -1.000 | 10.0 | 165.443858 | 183.038525 | 357.247596 | 407.714768 | 367.061131 | 403.271235 | 439.417678 | 457.963162 | ... | 191.327237 | 191.225602 | 181.691803 | 208.996170 | 198.116414 | 177.822697 | 192.377679 | 169.344080 | 137.058994 | 431.558397 |
| 1 | -0.996 | 10.0 | 169.920365 | 188.051328 | 368.793126 | 421.133352 | 377.604580 | 416.300607 | 454.302290 | 473.374693 | ... | 197.382219 | 197.558606 | 188.066255 | 215.492143 | 204.044079 | 183.797165 | 198.590959 | 175.259107 | 142.024406 | 445.426355 |
| 2 | -0.992 | 10.0 | 174.239568 | 192.901037 | 380.086587 | 434.450949 | 387.836302 | 429.071681 | 468.968133 | 488.561540 | ... | 203.319116 | 203.775189 | 194.335510 | 221.796086 | 209.799195 | 189.672437 | 204.685274 | 181.083009 | 146.922135 | 459.071033 |
| 3 | -0.988 | 10.0 | 178.391438 | 197.575344 | 391.093778 | 447.647728 | 397.730075 | 441.553881 | 483.380503 | 503.488717 | ... | 209.123305 | 209.858019 | 200.480026 | 227.885826 | 215.364812 | 195.433170 | 210.645100 | 186.798909 | 151.736835 | 472.466480 |
| 4 | -0.984 | 10.0 | 182.367373 | 202.063155 | 401.782197 | 460.704946 | 407.262053 | 453.719253 | 497.506821 | 518.123475 | ... | 214.781062 | 215.790568 | 206.480974 | 233.740487 | 220.725338 | 201.064896 | 216.455858 | 192.390601 | 156.453546 | 485.589296 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 50095 | 0.984 | 20.0 | 61.349063 | 65.048047 | 85.063819 | 113.433422 | 88.394031 | 124.506086 | 149.952411 | 121.336701 | ... | 42.527688 | 66.486529 | 41.358480 | 43.371996 | 44.445922 | 38.431979 | 55.741899 | 38.668228 | 32.639920 | 133.629536 |
| 50096 | 0.988 | 20.0 | 62.286521 | 66.031386 | 87.079793 | 115.201462 | 89.856116 | 126.484048 | 152.059416 | 121.359426 | ... | 42.937436 | 67.461370 | 41.915039 | 43.619696 | 45.178900 | 38.646578 | 56.871620 | 39.109777 | 32.910099 | 133.434442 |
| 50097 | 0.992 | 20.0 | 62.730903 | 66.501097 | 88.450080 | 116.113594 | 90.658314 | 127.473423 | 152.945087 | 120.539369 | ... | 43.032323 | 67.901196 | 42.151561 | 43.561366 | 45.569074 | 38.588749 | 57.545199 | 39.264615 | 32.936681 | 132.337261 |
| 50098 | 0.996 | 20.0 | 62.651405 | 66.423002 | 89.105845 | 116.103670 | 90.741003 | 127.405634 | 152.538994 | 118.845436 | ... | 42.792571 | 67.772755 | 42.045046 | 43.179045 | 45.587985 | 38.243719 | 57.726269 | 39.112192 | 32.705906 | 130.307051 |
| 50099 | 1.000 | 20.0 | 62.032828 | 65.779450 | 88.999047 | 115.132624 | 90.066964 | 126.244182 | 150.810430 | 116.272835 | ... | 42.208679 | 67.060129 | 41.583470 | 42.464753 | 45.218892 | 37.605378 | 57.393721 | 38.641578 | 32.211962 | 127.340661 |
50100 rows × 259 columns
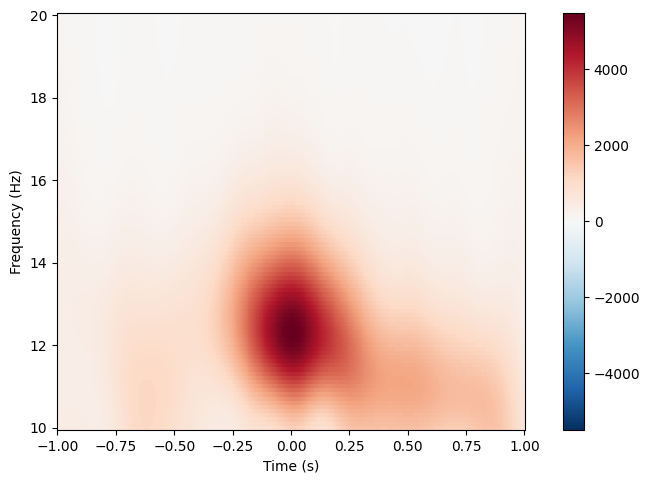
spindles_pipe.tfrs["N2"].plot(["E101"]),
spindles_pipe.tfrs["N3"].plot(["E101"])
No baseline correction applied
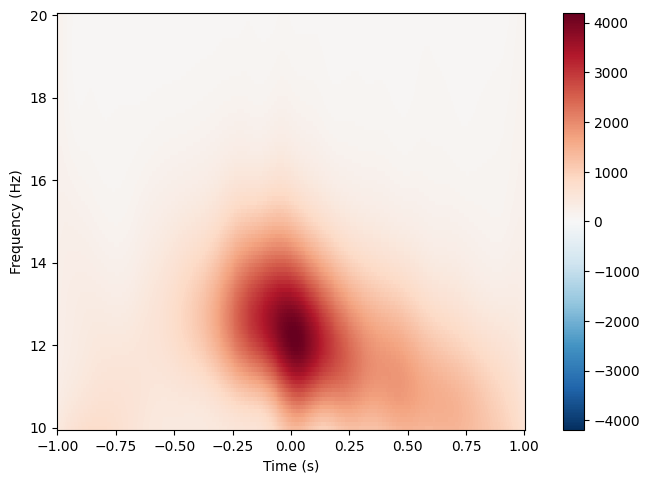
No baseline correction applied
[<Figure size 640x480 with 2 Axes>]
If you want to load saved TFR files:
spindles_pipe.read_tfrs(dirpath=None)