Spectral analyses
Introductory notes:
This notebook presents spectral analyses functionality:
Power spectral density (PSD) per sleep stage
Spectrogram & hypnogram
Topomaps for the spectra per sleep stage
Spectral parametrization with FOOOF
Additional results such as sleep statistics.
Recommended watching and reading:
Import data
Import module
from sleepeegpy.pipeline import SpectralPipe
from os import makedirs, path
import mne
mne.set_log_level("ERROR")
By default, all the input files are assumed to be saved in input_files, which will be created (if not already exists) in the notebook path. Change the following string to use another path
output_dir = "output_folder" # Output path and name can be changed here
input_dir = "input_files" # input files dir can be changed here
makedirs(input_dir, exist_ok=True)
makedirs(output_dir, exist_ok=True)
Add required files
Put all your files in the input folder.
Modify your eeg file name below. The file can be any format supported by the mne.read_raw() function.
Modify your hypnogram file name (Point-per-row type of hypnogram) below.
If needed, change Hypnogram’s sampling frequency
For more information about the supported formats, see mne documentation
eeg_file_name = "cleaned_raw.fif" # add your eeg_path here
hypnogram_filename = "staging.txt" # Point-per-row type of hypnogram.
hypno_freq = 1 # If required, change Hypnogram's sampling frequency (visbrain's hypnograms default to 1)
Initialize SpectralPipe object
path_to_eeg = path.join(input_dir, eeg_file_name)
hypnogram_path = path.join(input_dir, hypnogram_filename)
assert path.isfile(path_to_eeg) and path.isfile(
hypnogram_path
), f"{path_to_eeg} or {hypnogram_path} not exist"
spectral_pipe = SpectralPipe(
path_to_eeg=path_to_eeg,
output_dir=output_dir,
path_to_hypno=hypnogram_path,
hypno_freq=hypno_freq,
)
Hypnogram is SHORTER than data by 0.43 seconds. Padding hypnogram with last value to match data.size.
Compute PSD
spectral_pipe.compute_psd(
# A dict describing stages and their indices in the hypnogram file.
sleep_stages={"Wake": 0, "N1": 1, "N2": 2, "N3": 3, "REM": 4},
# Rereferencing to apply. Can be list of str channels or "average".
# If None, will not change the reference.
reference="average",
fmin=0, # Lower frequency bound.
fmax=60, # Upper frequency bound.
picks="eeg", # Channels to compute the PSD for.
reject_by_annotation=True, # Whether to reject epochs annotated as BAD.
save=True, # Whether to save the PSD hdf5 file for each sleep stage.
overwrite=True, # Whether to overwrite hdf5 files if there are any.
# Additional arguments passed to the PSD computing method, i.e., welch or multitaper:
n_fft=1024,
n_per_seg=1024,
n_overlap=512,
window="hamming",
n_jobs=-1,
verbose=False,
)
spectral_pipe.psds["REM"].get_data()
array([[1.20544608e-10, 9.13856487e-10, 4.32137885e-10, ...,
2.00851069e-14, 2.02085506e-14, 1.96803265e-14],
[9.16359851e-11, 6.92785547e-10, 3.43649234e-10, ...,
1.07580601e-14, 1.03970152e-14, 1.02861636e-14],
[4.65226376e-11, 3.45777189e-10, 1.85735238e-10, ...,
1.18917446e-14, 1.20680226e-14, 1.20086151e-14],
...,
[2.18358859e-11, 1.40802821e-10, 8.09673291e-11, ...,
1.62120327e-14, 1.51799036e-14, 1.53065000e-14],
[9.57268940e-12, 6.17214444e-11, 3.82661443e-11, ...,
1.17953057e-14, 1.11716153e-14, 1.09855943e-14],
[1.02225625e-11, 6.66994068e-11, 5.10107331e-11, ...,
3.09470840e-14, 3.19075520e-14, 3.06672304e-14]])
spectral_pipe.psds["REM"].to_data_frame()
| freq | E1 | E2 | E3 | E4 | E5 | E6 | E7 | E8 | E9 | ... | E248 | E249 | E250 | E251 | E252 | E253 | E254 | E255 | E256 | VREF | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.000000 | 1.205446e-10 | 9.163599e-11 | 4.652264e-11 | 2.361730e-11 | 1.874374e-11 | 1.450813e-11 | 2.218452e-11 | 1.122231e-11 | 9.538333e-12 | ... | 1.817369e-10 | 6.496992e-11 | 2.793785e-11 | 2.312580e-11 | 2.281494e-10 | 6.776294e-11 | 4.518227e-11 | 2.183589e-11 | 9.572689e-12 | 1.022256e-11 |
| 1 | 0.244141 | 9.138565e-10 | 6.927855e-10 | 3.457772e-10 | 1.662492e-10 | 1.119225e-10 | 8.076044e-11 | 9.857186e-11 | 6.725583e-11 | 5.687559e-11 | ... | 1.323533e-09 | 4.610354e-10 | 1.892631e-10 | 1.584058e-10 | 1.669154e-09 | 5.028152e-10 | 3.291209e-10 | 1.408028e-10 | 6.172144e-11 | 6.669941e-11 |
| 2 | 0.488281 | 4.321379e-10 | 3.436492e-10 | 1.857352e-10 | 1.034098e-10 | 7.332040e-11 | 5.620539e-11 | 5.915940e-11 | 5.096347e-11 | 4.554884e-11 | ... | 6.524901e-10 | 2.370033e-10 | 1.048493e-10 | 8.556182e-11 | 8.178159e-10 | 2.516436e-10 | 1.669304e-10 | 8.096733e-11 | 3.826614e-11 | 5.101073e-11 |
| 3 | 0.732422 | 1.305058e-10 | 1.122229e-10 | 6.907628e-11 | 4.777266e-11 | 3.865387e-11 | 3.290459e-11 | 3.159175e-11 | 3.215459e-11 | 2.766479e-11 | ... | 2.076041e-10 | 8.276927e-11 | 4.225638e-11 | 3.487731e-11 | 2.510315e-10 | 8.321513e-11 | 6.082972e-11 | 3.462790e-11 | 1.804304e-11 | 3.009207e-11 |
| 4 | 0.976562 | 5.479270e-11 | 4.879642e-11 | 3.398966e-11 | 2.742932e-11 | 2.447354e-11 | 2.202644e-11 | 2.154214e-11 | 2.232954e-11 | 1.853072e-11 | ... | 8.822444e-11 | 3.922727e-11 | 2.295007e-11 | 2.046545e-11 | 1.020850e-10 | 3.707458e-11 | 2.900152e-11 | 1.940715e-11 | 1.125545e-11 | 1.945493e-11 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 241 | 58.837891 | 2.052002e-14 | 1.080107e-14 | 1.185645e-14 | 9.236874e-15 | 9.386774e-15 | 9.163107e-15 | 1.086743e-14 | 1.460974e-14 | 1.055531e-14 | ... | 2.084268e-14 | 2.363470e-14 | 1.887338e-14 | 2.243271e-14 | 1.669183e-14 | 1.151799e-14 | 1.420598e-14 | 1.654172e-14 | 1.119376e-14 | 3.135501e-14 |
| 242 | 59.082031 | 2.008716e-14 | 1.057612e-14 | 1.171000e-14 | 9.409765e-15 | 8.857569e-15 | 9.199416e-15 | 1.060803e-14 | 1.466554e-14 | 9.855808e-15 | ... | 2.169877e-14 | 2.232925e-14 | 1.901087e-14 | 2.242874e-14 | 1.711121e-14 | 1.185718e-14 | 1.410618e-14 | 1.592892e-14 | 1.118600e-14 | 2.988033e-14 |
| 243 | 59.326172 | 2.008511e-14 | 1.075806e-14 | 1.189174e-14 | 9.523270e-15 | 8.751198e-15 | 9.190660e-15 | 1.006159e-14 | 1.388243e-14 | 9.746180e-15 | ... | 2.054704e-14 | 2.285352e-14 | 1.953269e-14 | 2.312350e-14 | 1.555995e-14 | 1.160546e-14 | 1.403677e-14 | 1.621203e-14 | 1.179531e-14 | 3.094708e-14 |
| 244 | 59.570312 | 2.020855e-14 | 1.039702e-14 | 1.206802e-14 | 9.175959e-15 | 8.892312e-15 | 8.748358e-15 | 1.008715e-14 | 1.370326e-14 | 9.504832e-15 | ... | 2.022414e-14 | 2.315599e-14 | 1.872794e-14 | 2.186576e-14 | 1.540562e-14 | 1.114618e-14 | 1.346594e-14 | 1.517990e-14 | 1.117162e-14 | 3.190755e-14 |
| 245 | 59.814453 | 1.968033e-14 | 1.028616e-14 | 1.200862e-14 | 9.139879e-15 | 8.727308e-15 | 8.709531e-15 | 1.034216e-14 | 1.343112e-14 | 9.215188e-15 | ... | 2.012729e-14 | 2.342522e-14 | 1.859850e-14 | 2.136766e-14 | 1.595666e-14 | 1.116042e-14 | 1.342614e-14 | 1.530650e-14 | 1.098559e-14 | 3.066723e-14 |
246 rows × 258 columns
Visualize
PSD
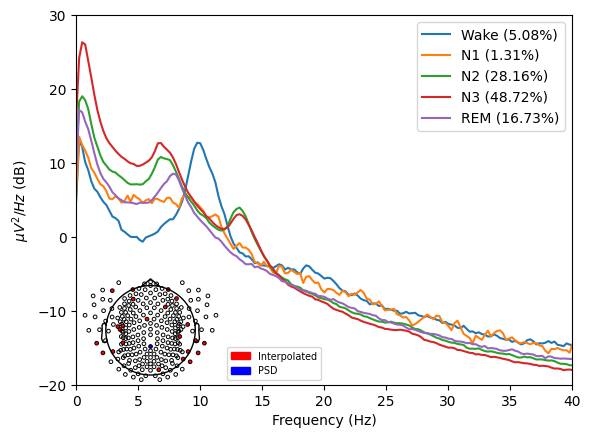
spectral_pipe.plot_psds(
picks=["E101"],
psd_range=(-20, 30), # Y axis limits
freq_range=(0, 40), # X axis limits
dB=True,
xscale="linear", # Matplotlib xscale. Can be {"linear", "log", "symlog", "logit", ...} or ScaleBase
axis=None,
plot_sensors=True, # Whether to plot EEG sensors showing which channels were used to compute PSD.
save=True, # Whether to save the plot as a png file.
)
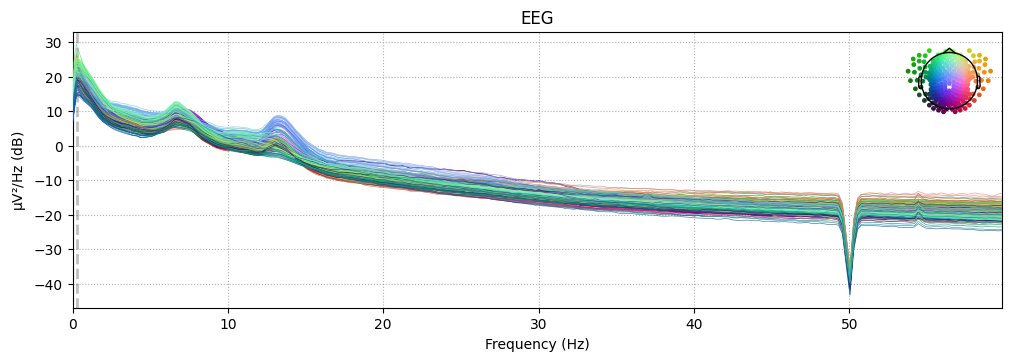
_ = spectral_pipe.psds["N2"].plot(picks="data", exclude="bads", show=False)
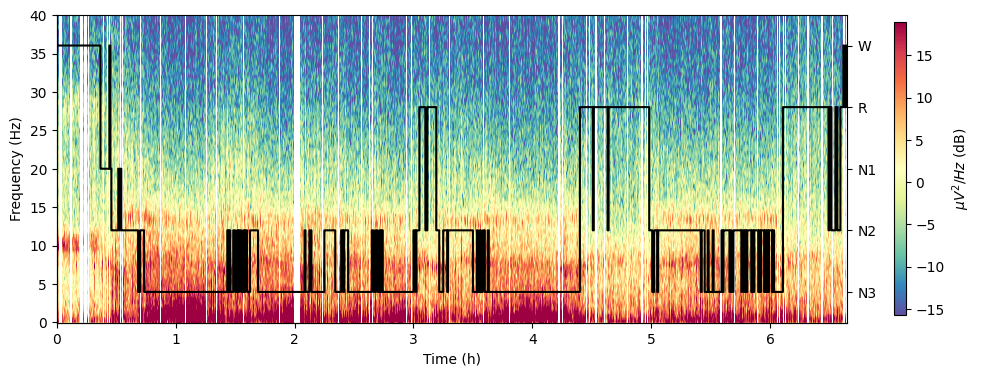
Hypnogram & spectrogram
spectral_pipe.plot_hypnospectrogram(
picks=["E101"], # Channel[s] to compute the spectrogram on.
win_sec=10, # The length of the sliding window, in seconds, used for multitaper PSD computation.
freq_range=(0, 40), # Y axis limits
cmap="Spectral_r", # Matplotlib colormap as in https://matplotlib.org/stable/tutorials/colors/colormaps.html
overlap=True, # Whether to plot hypnogram over spectrogram (True) or on top of it (False)
save=True, # Whether to save the plot as a file.
)
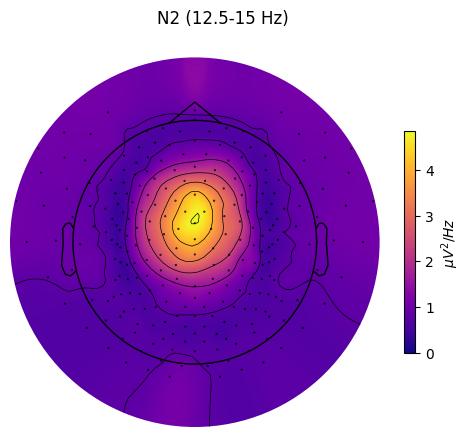
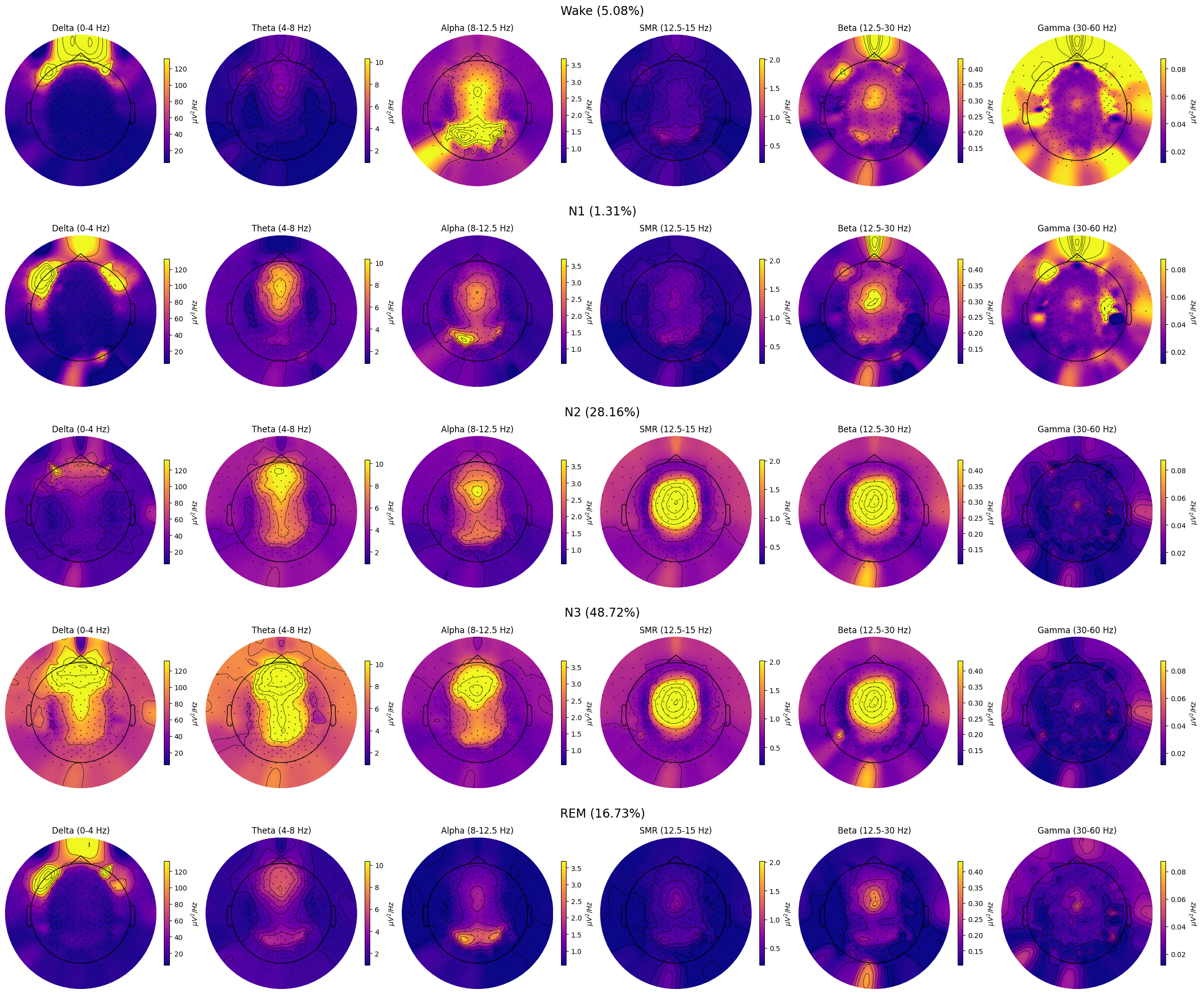
Topomap
Plots a topomap for a single sleep stage and frequency band
spectral_pipe.plot_topomap(
stage="N2", # Stage to plot topomap for.
band={"SMR": (12.5, 15)}, # Band to plot topomap for.
# Should contain at least index of the provided "stage".
dB=False, # Whether to transform PSD to dB/Hz
axis=None, # Whether to plot on provided matplotlib axis.
save=True, # Whether to save the plot as a file.
topomap_args=dict(cmap="plasma"), # Arguments passed to mne.viz.plot_topomap().
cbar_args=None, # Arguments passed to plt.colorbar().
)
Topomap collage
Plot topomaps for multiple bands and sleep stages
spectral_pipe.plot_topomap_collage(
# Bands to plot topomaps for.
bands={
"Delta": (0, 4),
"Theta": (4, 8),
"Alpha": (8, 12.5),
"SMR": (12.5, 15),
"Beta": (12.5, 30),
"Gamma": (30, 60),
},
# Tuple of strs or "all", e.g., ("N1", "REM") or "all" (plots all "sleep_stages").
stages_to_plot="all",
dB=False, # Whether to transform PSD to dB/Hz.
low_percentile=5, # Set min color value by percentile of the band data.
high_percentile=95, # Set max color value by percentile of the band data.
fig=None, # Instance of plt.Figure, a new fig will be created if None.
save=True, # Whether to save the plot as a file.
topomap_args=dict(cmap="plasma"), # Arguments passed to mne.viz.plot_topomap().
cbar_args=None, # Arguments passed to plt.colorbar().
)
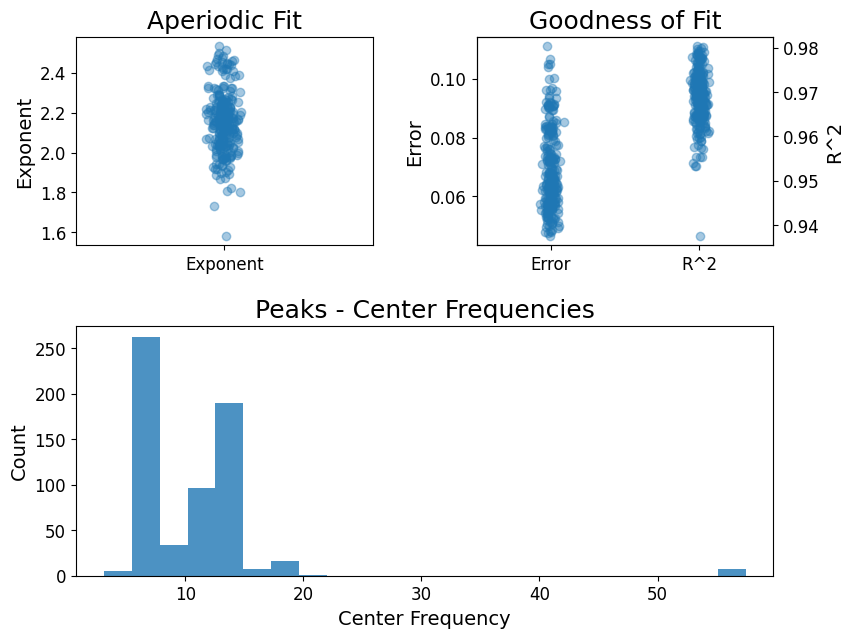
Parametrize spectrum
spectral_pipe.parametrize(
picks=["eeg"], # Channels to use.
freq_range=[0.5, 60], # Range of frequencies to parametrize.
# Whether to average psds over channels.
# If False or multiple channels are provided, the FOOOFGroup will be used.
# Defaults to False.
average_ch=False,
)
spectral_pipe.fooofs["N2"].report()
Running FOOOFGroup across 257 power spectra.
==================================================================================================
FOOOF - GROUP RESULTS
Number of power spectra in the Group: 257
The model was run on the frequency range 0 - 60 Hz
Frequency Resolution is 0.24 Hz
Power spectra were fit without a knee.
Aperiodic Fit Values:
Exponents - Min: 1.583, Max: 2.533, Mean: 2.158
In total 618 peaks were extracted from the group
Goodness of fit metrics:
R2s - Min: 0.938, Max: 0.980, Mean: 0.969
Errors - Min: 0.047, Max: 0.111, Mean: 0.069
==================================================================================================
Sleep Stats
spectral_pipe.sleep_stats(save=False)
{'TIB': 399.0405333333333,
'SPT': 376.5,
'WASO': 1.0,
'TST': 375.5,
'N1': 6.0,
'N2': 109.5,
'N3': 191.0,
'REM': 69.0,
'NREM': 306.5,
'SOL': 22.0,
'Lat_N1': 22.0,
'Lat_N2': 27.5,
'Lat_N3': 41.0,
'Lat_REM': 183.0,
'%N1': 1.5978695073235685,
'%N2': 29.161118508655125,
'%N3': 50.865512649800266,
'%REM': 18.37549933422104,
'%NREM': 81.62450066577897,
'SE': 94.10071625138166,
'SME': 99.734395750332}